Creating a bee-specific database
Arguably, 454 pyrosequencing has revolutionized the field of
microbial ecology. Where it was once costly to generate libraries of a
few hundred 16S rRNA gene sequences, 454 pyrosequencing allows researchers to
deeply probe a microbial community at relatively little cost per
sequence. The ultimate goal of 454 pyrosequencing amplicon studies is to
characterize a microbial community, either in terms of composition (DNA) or
activity (RNA). A large number of groups have been using the Ribosomal Database
Project's Naïve Bayesian Classifier (RDP-NBC) to achieve this goal (Wang et
al., 2007). The advantages are numerous but I'll list a few of the practical
ones here: classification is straightforward (putting sequences in their
taxonomic context), efficient (especially when considering tens of thousands of
sequences) and does not require full length 16S sequences (making it an
appropriate tool for pyrosequencing studies). However, the NBC relies on
an accurate training set – on reference sequences used to train the model and
generate the classification results. In a publication by Werner et al.
(2011), the training set had a significant impact on classification, improving
the classification of previously “unclassified” sequences and increasing the
number classified to genus [1].
For environments that lack cultured isolates or are relatively
unexplored, it can be difficult to find the appropriate training set to reveal
the true taxonomic identity of the sequences extracted. However, if
previous clone libraries have generated full length, high-quality 16S sequences
these can be added to the seed alignment and the taxonomy framework. This
is what I've aimed to do for the honey bee gut, using Mothur. In Mothur you can
“tweak” the alignment seed for any particular environment, creating a custom
database that will more accurately classify sequences of interest.
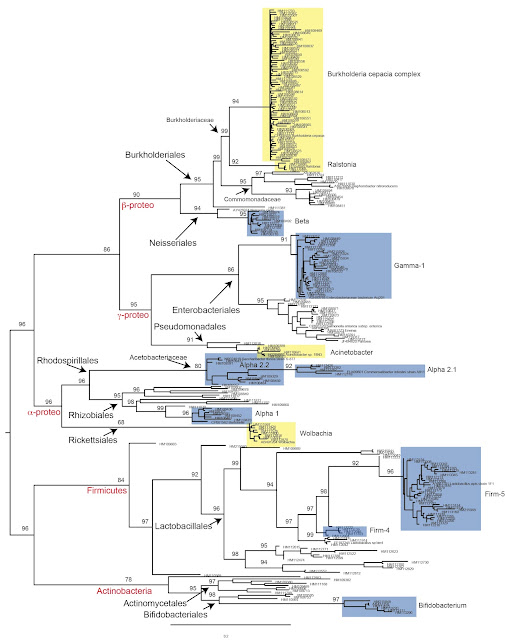
To create a bee-specific alignment compatible with Mothur you need
two files: a reference database and a taxonomy file for each of those
sequences. To generate the database I downloaded all sequences that
corresponded to accession numbers published in analyses of bee-associated
microbiota and that were near full length (1250 bp) (A total of 5,713 sequences
were downloaded and 5,158 passed the length threshold). These
sequences were clustered at 99% identity, reducing the dataset to 276
representatives. This set of sequences were aligned using the SINA
aligner (v 1.2.9, [2]) to the arb-silva SSU database
(SSURef_108_SILVA_NR_99_11_10_11_opt_v2.arb) and visually inspected using ARB [3]. To generate a phylogeny I used this
aligned sequence set as input to RAxML (GTR+g with 1000 bootstrap replicates)
using a maximum likelihood framework [4]. Taking a
quick look at this taxonomy, it is clear that we've got the majority of
sequences falling into either the Firmicutes, Actinobacteria, or Proteobacteria.
Also, specific clades identified by previous groups are clearly marked on
the tree (based on the literature). Now, it's difficult to classify novel
sequences so you might be asking your self, how could you taxonomically
classify them based on this tree? Fine scale taxonomic placement (below
phylum level) for relatively novel bacterial groups is difficult to accomplish
and subject to some debate [5]. So, I queried the RDP for nearest
cultured representatives. If these cultured representative was
>95% identical to the bee derived sequence then that novel sequence was
placed in the genus of the cultured representative. If, however, the sequence
identity above 95% was not found for these sequences in the cultured isolates,
but they claded with a cultured representative, they were placed in the same
phylum, class, or order (depending on the group) and we noted incerte
sedis in the taxonomy file. In addition to this de
novo generation of taxonomic information for these bee sequences, if
phylogenetic information (as established by Cox-Foster et al., 2006)
was associated with any of these Genbank submissions, that information was also
included in the taxonomy.
I then downloaded each of three pre-existing,
Mothur-compatible training sets: 1) the RDP 16S rRNA reference v7 (9,662
sequences), 2) the Greengenes reference (84,414 sequences), and 3) the SILVA
bacterial reference (14,956 sequences) each available on the Mothur WIKI page
(http://www.mothur.org/wiki/Main_Page). These datasets are comprised of
both an unaligned sequence file and a taxonomy file. To each of these I
added the honey bee specific training set I generated. Using each of these
six alternative datasets (either with or without the honey bee specific
sequences), I classified the honey bee gut microbiota generated in our
recent publication [4] using the RDP-II Naive Bayesian
Classifier [6] and a 60%
confidence threshold.
What I find most interesting about this analysis is how well the
addition of the bee-specific sequences helps to create congruence among the
datasets (the Orbus classification by RDP not withstanding). Clearly, inclusion of environment specific
sequences can increase the accuracy of the RDP-NBC. I wanted to use this framework to explore
fine-scale diversity (OTU level) within the gut.
Figure 2. The effect of training set on the classification of sequences from the
honey bee gut visualized by a heat map.
Unique sequences (4,480) were classified using the NBC trained on either
RDP, GG, or SILVA (A) or three custom databases including near full length
honey bee-associated sequences RDP+bees, GG+bees, SILVA+bees (B). The effect of including custom sequences is
most obvious in the classification discordance between RDP, GG and SILVA and
their relative congruence when honey bee associated sequences are added to the
training set (B).
Below I ask, how many individual unique
sequences and how many likely "species" do we find in each of these families
based on 97% clustering of operational taxonomic units (OTUs)? (Table 1).
Table 1. For
each family found with honey bee guts, the number of unique sequences and the
number of 97% identity operational taxonomic units (OTUs) is shown. The taxonomy shown here is based on
classification by the RDP-NBC using the SILVA + honey bee sequences training
set (available for anyone via email). The
most abundant families represent a large amount of fine-scale bacterial diversity.
Family
|
Num. unique sequences
|
OTUs
|
Enterobacteriaceae
|
1621
|
175
|
gamma-1
|
436
|
48
|
beta
|
532
|
35
|
Bifidobacteriaceae
|
363
|
32
|
firm-5
|
929
|
32
|
firm-4
|
253
|
21
|
alpha-2.1
|
90
|
15
|
alpha-1
|
65
|
13
|
Lactobacilliaceae
|
86
|
12
|
Flavobacteriaceae
|
2
|
2
|
Leuconostocaceae
|
2
|
2
|
Moraxellaceae
|
6
|
2
|
Sphingomonadaceae
|
2
|
2
|
Xanthomonadaceae
|
2
|
2
|
Actinomycetaceae
|
1
|
1
|
Aeromonadaceae
|
1
|
1
|
alpha-2.2
|
10
|
1
|
Clostridiaceae
|
2
|
1
|
Corynebacteriaceae
|
1
|
1
|
Cytophagaceae
|
1
|
1
|
Enterococcaceae
|
9
|
1
|
Incertae_Sedis_XI
|
1
|
1
|
Kineosporiaceae
|
1
|
1
|
Nakamurellaceae
|
1
|
1
|
Oxalobacteraceae
|
1
|
1
|
Prevotellaceae
|
1
|
1
|
One central goal of our previous study (see [7]) was to determine if there was a difference between colonies generated from promiscuous honey bee queens and those that were relatively chaste. When we compared the OTU content between these two colony types, we found that the genetically diverse colonies host more diverse microflora [7]. This difference, based on number of 97% identity clusters found within each colony, is independent of classification and was recapitulated using the SILVA + honey bee taxonomic classification (the 95% confidence interval (CI) for mean difference between species diversity compared between colony types exceeded 0; 95% CI = 110, 102; mean = 106.25).
The next question is, is
this difference in microbiota composition attributable to any specific
taxonomic group? That is, within
specific bacterial families, do we see differences between genetically diverse
and genetically uniform colonies with regards to their OTU content? I used the bootstrapped confidence interval
analysis used in [7] to answer this question. The difference in genetic diversity between
colony types was found to effect the OTU-level diversity of specific bacterial groups
(Table 2). This suggests that fine-scale diversity within these honey-bee
specific families may be ecologically relevant, and shouldn’t be ignored.
Table 2. Total
number of operational taxonomic units (97% ID) in either genetically uniform or
genetically diverse colonies and classified as one of the honey bee specific
taxonomic groups (mean number used in CI calculation in parentheses). Statistically significant differences between
colony types was observed for most of these families and their OTU content
(indicated by an asterisk).
Taxon
|
Genetically Diverse
|
Genetically Uniform
|
Bootstrap 95% CI of mean
difference
|
Firm-4*
|
44 (36.10)
|
25 (25.04)
|
(11.22, 10.90)
|
Firm-5*
|
56 (45.13)
|
46 (46.05)
|
(0.74,1.09)
|
Alpha-2.1*
|
21(16.03)
|
21 (21.04)
|
(4.92, 5.09)
|
Alpha-2.2
|
4 (4.05)
|
4 (4.01)
|
(0.09, -0.005)
|
Alpha-1*
|
16 (12.01)
|
13 (13.06)
|
(0.96, 1.14)
|
Beta*
|
60 (48.98)
|
38 (37.99)
|
(11.12, 10.85)
|
Gamma-1*
|
66 (52.73)
|
51 (50.99)
|
(1.94, 1.55)
|
Which brings me to a final
point – that’s more of a rant really.
This is about lumping and splitting – you say “toma-toe” and I say
“tomah-toe” – and what one calls a bacterial “species” (I’m not stepping into
that mine field). We don’t yet know what the observed %
divergence at the 16S rRNA gene means in the honey bee gut microbiota. Could these differences be primarily
attributable to diversity between operons within a single strain? This is unlikely; within the majority of bacterial
genomes these operons evolve by concerted evolution and show <1% divergence
between gene copies [8]. We are using
the 16S rRNA gene as a marker for diversity – as a taxonomic tag. This short tag is just that – a marker. It could represent an enormous diversity at
the genome level, we don’t yet know. What
we do know is that the microbial world is vast, that diversity is the norm –
very few environments are characterized by low species abundance or “clonal”
strains. The fact that we are able to
pick up a statistically significant signal between honey bee colonies based on OTUs
suggests to me that there is more to investigate here.


Comments
Post a Comment